Photo by Janna Nichols
SeaDoc Society is excited to fund a project to help and save the Salish Sea’s only abalone species through a new technique involving DNA
Pinto abalone populations in the Salish Sea have been decimated after years of poaching and over-harvest, falling by 98 percent in recent decades. There are not enough remaining in the wild for them to reproduce naturally because they are “broadcast spawners,” meaning if one releases sperm, a female needs to release her eggs close enough to be fertilized.
When abalone are not close enough to other reproductively active abalone there can be no reproduction, making them functionally extinct in the wild.
SeaDoc-funded studies have been crucial for providing data needed to recover pinto abalone, from genetic work to assessment of habitat to optimization of releasing captively bred animals.
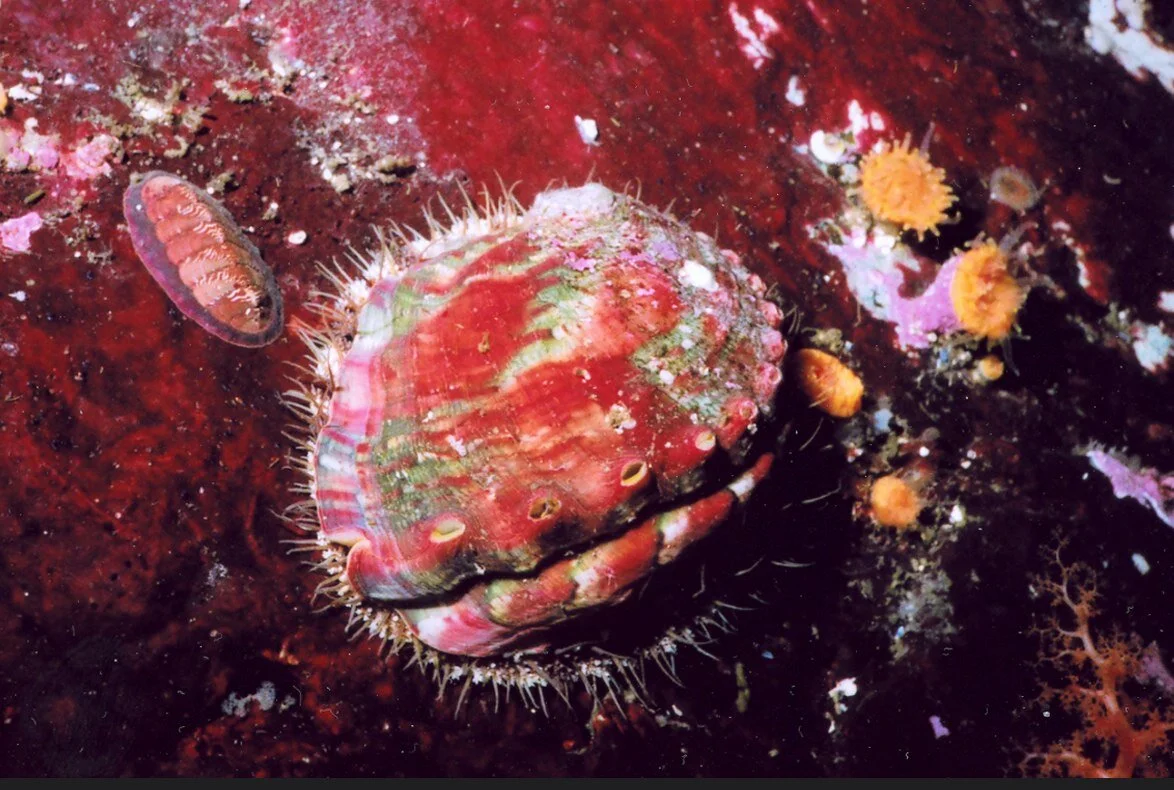
Hatchery-reared Pinto abalone performs escape mechanism
Each year, we fund several important research projects with an eye toward improving policy and conservation. We’re thrilled to announce the first of our funded projects for 2020, which could optimize the way we identify wild habitat for abalone recovery.
A major hurdle in saving the species is the lack of an efficient, cost-effective way to detect where they live and thrive throughout the Salish Sea, which brings us to an exciting project being led by Dr. James Dimond of Western Washington University.
Bottling up abalone DNA
The current method for detecting abalone populations is to take a dive crew out on a boat and send them down in search of these cryptic endangered snails. Fun? Of course! But it’s also laborious and expensive, which puts a pretty hard limit on how much ground a dive team can efficiently cover.
Dimond’s team will test a method that involves collecting water from multiple locations and sampling the water for abalone DNA to determine if abalone are present.
“We’ll take a two-liter Nalgene bottle, dip it in the water and filter the water with a vacuum pump,” said Dimond. “The water runs through a fine filter that will retain the DNA, which is then preserved for later analysis.”
Once the team has collected all of the samples, they take the preserved filters, grind them up, and extract the DNA. There will obviously be more than just abalone in those samples, but their test will measure for the specific mitochondrial DNA of pinto abalone—sort of like a pinto abalone bar code.
How they’ll do it
The first phase will test the method in an aquarium setting, where Dimond and his team can be certain which samples should and should not test positive for pinto abalone.
In phase two they’ll bring the method to the sea, starting at an abalone restoration site, where they’ll gather Nalgene bottle samples from two meters away, five meters away, and the sea surface. They’ll measure the results at successive distances from abalone aggregations to test and determine the method’s effectiveness.
The final phase will be to test the method in a truly wild setting to see if they can locate areas within the Salish Sea with high densities of pinto abalone, which would yield valuable information for the biologists who are breeding abalone in captivity to release this endangered species back into the wild.
“I’m aiming to sample 50 sites in five or six days,” said Dimond. “To do that with divers would take weeks and you’d need a full team. We can do this with a skipper and two people.”
SeaDoc Science Director Joe Gaydos added:
“This is an example of how cutting-edge science can be used to help recover and monitor endangered species,” he said. “I think we could bring back pinto abalone in the next decade and this could be very important for tracking their recovery.”